All of the Following Can Form Filaments Except __________.
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Businesswoman S, editor. Medical Microbiology. 4th edition. Galveston (TX): University of Texas Medical Branch at Galveston; 1996.

General Concepts
Gross Morphology
Bacteria have characteristic shapes (cocci, rods, spirals, etc.) and ofttimes occur in characteristic aggregates (pairs, chains, tetrads, clusters, etc.). These traits are usually typical for a genus and are diagnostically useful.
Jail cell Structure
Prokaryotes accept a nucleoid (nuclear body) rather than an enveloped nucleus and lack membrane-bound cytoplasmic organelles. The plasma membrane in prokaryotes performs many of the functions carried out by bleary organelles in eukaryotes. Multiplication is by binary fission.
Surface Structures
Flagella: The flagella of motile leaner differ in structure from eukaryotic flagella. A basal body anchored in the plasma membrane and cell wall gives rise to a cylindrical protein filament. The flagellum moves by whirling virtually its long axis. The number and system of flagella on the cell are diagnostically useful.
Pili (Fimbriae): Pili are slender, hairlike, proteinaceous appendages on the surface of many (particularly Gram-negative) bacteria. They are important in adhesion to host surfaces.
Capsules: Some leaner form a thick outer sheathing of high-molecular-weight, gluey polysaccharide gel; others have more amorphous slime layers. Capsules confer resistance to phagocytosis.
Important Chemical Components of Surface Structures
Cell Wall Peptidoglycans: Both Gram-positive and Gram-negative bacteria possess cell wall peptidoglycans, which confer the characteristic cell shape and provide the cell with mechanical protection. Peptidoglycans are unique to prokaryotic organisms and consist of a glycan backbone of muramic acid and glucosamine (both Due north-acetylated), and peptide chains highly cross-linked with bridges in Gram-positive leaner (eastward.m., Staphylococcus aureus) or partially cross-linked in Gram-negative bacteria (e.g., Escherichia coli). The cross-linking transpeptidase enzymes are some of the targets for b-lactam antibiotics.
Teichoic Acids: Teichoic acids are polyol phosphate polymers bearing a strong negative charge. They are covalently linked to the peptidoglycan in some Gram-positive bacteria. They are strongly antigenic, merely are mostly absent in Gram-negative bacteria.
Lipoteichoic Acids: Lipoteichoic acids as membrane teichoic acids are polymers of amphiphitic glycophosphates with the lipophilic glycolipid and anchored in the cytoplasmic membrane. They are antigenic, cytotoxic and adhesins (e.thousand., Streptococcus pyogenes).
Lipopolysaccharides: One of the major components of the outer membrane of Gram-negative bacteria is lipopolysaccharide (endotoxin), a complex molecule consisting of a lipid A anchor, a polysaccharide core, and chains of carbohydrates. Sugars in the polysaccharide chains confer serologic specificity.
Wall-Less Forms: Two groups of bacteria devoid of cell wall peptidoglycans are the Mycoplasma species, which possess a surface membrane structure, and the L-forms that arise from either Gram-positive or Gram-negative bacterial cells that have lost their ability to produce the peptidoglycan structures.
Cytoplasmic Structures
Plasma Membrane: The bacterial plasma membrane is composed primarily of poly peptide and phospholipid (about iii:1). It performs many functions, including transport, biosynthesis, and energy transduction.
Organelles: The bacterial cytoplasm is densely packed with 70S ribosomes. Other granules stand for metabolic reserves (due east.g., poly-β-hydroxybutyrate, polysaccharide, polymetaphosphate, and metachromatic granules).
Endospores: Bacillus and Clostridium species can produce endospores: rut-resistant, dehydrated resting cells that are formed intracellularly and contain a genome and all essential metabolic machinery. The endospore is encased in a circuitous protective spore coat.
Introduction
All bacteria, both pathogenic and saprophytic, are unicellular organisms that reproduce by binary fission. Nigh bacteria are capable of independent metabolic existence and growth, but species of Chlamydia and Rickettsia are obligately intracellular organisms. Bacterial cells are extremely small and are near conveniently measured in microns (10-half-dozen chiliad). They range in size from large cells such as Bacillus anthracis (one.0 to ane.3 µm Ten iii to 10 µm) to very small cells such every bit Pasteurella tularensis (0.two Ten 0.2 to 0.7 µm) Mycoplasmas (atypical pneumonia group) are even smaller, measuring 0.1 to 0.2 µm in diameter. Bacteria therefore accept a surface-to-book ratio that is very high: about 100,000.
Leaner have feature shapes. The common microscopic morphologies are cocci (round or ellipsoidal cells, such equally Staphylococcus aureus or Streptococcus, respectively); rods, such every bit Bacillus and Clostridium species; long, filamentous branched cells, such as Actinomyces species; and comma-shaped and spiral cells, such as Vibrio cholerae and Treponema pallidum, respectively. The arrangement of cells is also typical of diverse species or groups of leaner (Fig. 2-1). Some rods or cocci characteristically abound in chains; some, such as Staphylococcus aureus, form grapelike clusters of spherical cells; some round cocci grade cubic packets. Bacterial cells of other species grow separately. The microscopic advent is therefore valuable in classification and diagnosis. The higher resolving ability of the electron microscope not only magnifies the typical shape of a bacterial prison cell merely also conspicuously resolves its prokaryotic organisation (Fig. ii-ii).
Figure 2-ane
Typical shapes and arrangements of bacterial cells.
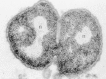
Effigy 2-2
Electron micrograph of a thin section of Neisseria gonorrhoeae showing the organizational features of prokaryotic cells. Note the electron-transparent nuclear region (n) packed with Deoxyribonucleic acid fibrils, the dumbo distribution of ribosomal particles in the cytoplasm, (more...)
The Nucleoid
Prokaryotic and eukaryotic cells were initially distinguished on the basis of structure: the prokaryotic nucleoidthe equivalent of the eukaryotic nucleusis structurally simpler than the true eukaryotic nucleus, which has a complex mitotic apparatus and surrounding nuclear membrane. As the electron micrograph in Fig. two-2 shows, the bacterial nucleoid, which contains the DNA fibrils, lacks a limiting membrane. Under the light microscope, the nucleoid of the bacterial cell can be visualized with the assist of Feulgen staining, which stains Deoxyribonucleic acid. Gentle lysis tin can be used to isolate the nucleoid of near bacterial cells. The Deoxyribonucleic acid is so seen to be a single, continuous, "giant" circular molecule with a molecular weight of approximately three X ten9 (see Ch. 5). The unfolded nuclear Deoxyribonucleic acid would be about ane mm long (compared with an average length of 1 to 2 µm for bacterial cells). The bacterial nucleoid, and then, is a structure containing a single chromosome. The number of copies of this chromosome in a prison cell depends on the stage of the cell bike (chromosome replication, cell enlargement, chromosome segregation, etc). Although the machinery of segregation of the ii sister chromosomes post-obit replication is non fully understood, all of the models proposed require that the chromosome be permanently attached to the cell membrane throughout the various stages of the cell cycle.
Bacterial chromatin does not incorporate basic histone proteins, but low-molecular-weight polyamines and magnesium ions may fulfill a function similar to that of eukaryotic histones. Despite the differences betwixt prokaryotic and eukaryotic DNA, prokaryotic Dna from cells infected with bacteriophage 𝛄, when visualized by electron microscopy, has a beaded, condensed advent not dissimilar that of eukaryotic chromatin.
Surface Appendages
2 types of surface appendage can exist recognized on certain bacterial species: the flagella, which are organs of locomotion, and pili (Latin hairs), which are also known as fimbriae (Latin fringes). Flagella occur on both Gram-positive and Gram-negative bacteria, and their presence can be useful in identification. For case, they are found on many species of bacilli simply rarely on cocci. In contrast, pili occur virtually exclusively on Gram-negative leaner and are plant on simply a few Gram-positive organisms (e.1000., Corynebacterium renale).
Some bacteria accept both flagella and pili. The electron micrograph in Fig. 2-three shows the characteristic wavy appearance of flagella and two types of pili on the surface of Escherichia coli.
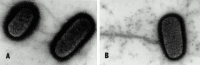
Figure 2-3
(A) Electron micrograph of negatively stained East. coli showing wavy flagella and numerous short, thinner, and more rigid hairlike structures, the pili. (B) The long sexual activity pilus can exist distinguished from the shorter mutual pili by mixing Due east. coli cells with (more...)
Flagella
Structurally, bacterial flagella are long (3 to 12 µm), filamentous surface appendages near 12 to thirty nm in diameter. The protein subunits of a flagellum are assembled to form a cylindrical structure with a hollow core. A flagellum consists of three parts: (i) the long filament, which lies external to the cell surface; (2) the hook structure at the end of the filament; and (3) the basal torso, to which the hook is anchored and which imparts motion to the flagellum. The basal torso traverses the outer wall and membrane structures. It consists of a rod and one or two pairs of discs. The thrust that propels the bacterial jail cell is provided past counterclockwise rotation of the basal body, which causes the helically twisted filament to whirl. The movement of the basal body is driven past a proton motive strength rather than by ATP direct. The ability of leaner to swim by means of the propeller-like action of the flagella provides them with the mechanical ways to perform chemotaxis (move in response to attractant and repellent substances in the surroundings). Response to chemical stimuli involves a sophisticated sensory system of receptors that are located in the cell surface and/or periplasm and that transmit information to methyl-accepting chemotaxis proteins that control the flagellar motor. Genetic studies have revealed the beingness of mutants with contradistinct biochemical pathways for flagellar motion and chemotaxis.
Chemically, flagella are constructed of a grade of proteins called flagellins. The hook and basal-body structures consist of numerous proteins. Mutations affecting whatsoever of these gene products may event in loss or harm of movement. Flagellins are immunogenic and constitute a grouping of protein antigens called the H antigens, which are characteristic of a given species, strain, or variant of an organism. The species specificity of the flagellins reflects differences in the master structures of the proteins. Antigenic changes of the flagella known as the phase variation of H1 and H2 occurs in Salmonella typhimurium (run into Ch. 21 and Ref. Seifert and then).
The number and distribution of flagella on the bacterial surface are characteristic for a given species and hence are useful in identifying and classifying bacteria. Figure ii-4 illustrates typical arrangements of flagella on or around the bacterial surface. For case, V. cholerae has a single flagellum at ane pole of the prison cell (i.e., it is monotrichous), whereas Proteus vulgaris and E. coli accept many flagella distributed over the entire cell surface (i.e., they are peritrichous). The flagella of a peritrichous bacterium must aggregate as a posterior bundle to propel the jail cell in a forward direction.

Effigy 2-4
Typical arrangements of bacterial flagella.
Flagella can exist sheared from the cell surface without affecting the viability of the prison cell. The cell and so becomes temporarily nonmotile. In time information technology synthesizes new flagella and regains motility. The poly peptide synthesis inhibitor chloramphenicol, even so, blocks regeneration of flagella.
Pili
The terms pili and fimbriae are usually used interchangeably to describe the thin, hairlike appendages on the surface of many Gram-negative bacteria and proteins of pili are referred to equally pilins. Pili are more than rigid in appearance than flagella (Fig. ii-3). In some organisms, such every bit Shigella species and E. coli, pili are distributed profusely over the cell surface, with equally many as 200 per cell. As is easily recognized in strains of Eastward. coli, pili can come in two types: short, arable common pili, and a small-scale number (ane to half dozen) of very long pili known as sex pili. Sex pili can exist distinguished by their power to bind male person-specific bacteriophages (the sex activity pilus acts as a specific receptor for these bacteriophages) (Fig. two-3B). The sex pili adhere male to female bacteria during conjugation.
Pili in many enteric bacteria confer adhesive properties on the bacterial cells, enabling them to adhere to various epithelial surfaces, to red blood cells (causing hemagglutination), and to surfaces of yeast and fungal cells. These adhesive properties of piliated cells play an of import role in bacterial colonization of epithelial surfaces and are therefore referred to as colonization factors. The common pili found on Due east. coli exhibit a saccharide specificity analogous to that of phytohemagglutinins and lectins, in that adhesion and hemagglutinating capacities of the organism are inhibited specifically by mannose. Organisms possessing this blazon of hemagglutination are called mannose-sensitive organisms. Other piliated organisms, such as gonococci, are agglutinative and hemagglutinating, but are insensitive to the inhibitory effects of mannose. Extensive antigenic variations in pilins of gonococci are well known (run across Ref. Seifert and So).
Surface Layers
The surface layers of the bacterial cell have been identified past diverse techniques: lite microscopy and staining; electron microscopy of thin-sectioned, freeze-fractured, and negatively stained cells; and isolation and biochemical label of individual morphologic components of the cell. The principal surface layers are capsules and loose slime, the cell wall of Gram-positive bacteria and the circuitous jail cell envelope of Gram-negative bacteria, plasma (cytoplasmic) membranes, and mesosomal membrane vesicles, which arise from invaginations of the plasma membrane. In bacteria, the prison cell wall forms a rigid construction of uniform thickness around the cell and is responsible for the characteristic shape of the cell (rod, coccus, or spiral). Inside the jail cell wall (or rigid peptidoglycan layer) is the plasma (cytoplasmic) membrane; this is unremarkably closely apposed to the wall layer. The topographic relationships of the cell wall and envelope layers to the plasma membrane are indicated in the thin section of a Gram-positive organism (Micrococcus lysodeikticus) in Figure ii-5A and in the freeze-fractured cell of a Gram-negative organism (Bacteroides melaninogenicus) in Figure two-5B. The latter shows the typical fracture planes seen in nearly Gram-negative bacteria, which are weak cleavage planes through the outer membrane of the envelope and extensive fracture planes through the bilayer region of the underlying plasma membrane.
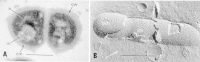
Figure 2-five
(A) Electron micrograph of a thin section of the Gram-positive M. lysodeikticus showing the thick peptidoglycan cell wall (cw), underlying cytoplasmic (plasma) membrane (cm), mesosome (m), and nucleus (n). (B) Freeze-fractured Bacteriodes cell showing (more...)
Capsules and Loose Slime
Some leaner course capsules, which constitute the outermost layer of the bacterial cell and surround it with a relatively thick layer of viscous gel. Capsules may be up to 10 µm thick. Some organisms lack a well-divers capsule simply accept loose, amorphous slime layers external to the jail cell wall or cell envelope. The α hemolytic Streptococcus mutans, the primary organism found in dental plaque is able to synthesis a large extracellular mucoid glucans from sucrose. Non all bacterial species produce capsules; however, the capsules of encapsulated pathogens are often important determinants of virulence. Encapsulated species are plant amid both Gram-positive and Gram-negative bacteria. In both groups, most capsules are composed of highmolecular-weight sticky polysaccharides that are retained equally a thick gel exterior the cell wall or envelope. The capsule of Bacillus anthracis (the causal agent of anthrax) is unusual in that it is composed of a γ-glutamyl polypeptide. Table two-1 presents the various capsular substances formed by a selection of Gram-positive and Gram-negative bacteria. A plasma membrane phase is involved in the biosynthesis and assembly of the capsular substances, which are extruded or secreted through the outer wall or envelope structures. Mutational loss of enzymes involved in the biosynthesis of the capsular polysaccharides tin event in the smooth-to-rough variation seen in the pneumococci.
Tabular array 2-1
Nature of Capsular Substances Formed by Various Bacteria.
The sheathing is non essential for viability. Viability is non affected when capsular polysaccharides are removed enzymatically from the cell surface. The exact functions of capsules are non fully understood, only they do confer resistance to phagocytosis and hence provide the bacterial cell with protection confronting host defenses to invasion.
Prison cell Wall and Gram-Negative Cell Envelope
The Gram stain broadly differentiates leaner into Gram-positive and Gram-negative groups; a few organisms are consistently Gram-variable. Gram-positive and Gram-negative organisms differ drastically in the arrangement of the structures outside the plasma membrane but below the capsule (Fig. two-half dozen): in Gram-negative organisms these structures constitute the jail cell envelope, whereas in Gram-positive organisms they are chosen a cell wall.
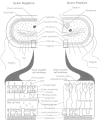
Figure two-six
Comparison of the thick cell wall of Gram-positive bacteria with the comparatively thin jail cell wall of Gram-negative bacteria. Note the complexity of the Gram-negative prison cell envelope (outer membrane, its hydrophobic lipoprotein anchor; periplasmic space). (more...)
Most Gram-positive bacteria have a relatively thick (nigh 20 to 80 nm), continuous jail cell wall (often called the sacculus), which is equanimous largely of peptidoglycan (also known every bit mucopeptide or murein). In thick cell walls, other cell wall polymers (such every bit the teichoic acids, polysaccharides, and peptidoglycolipids) are covalently fastened to the peptidoglycan. In contrast, the peptidoglycan layer in Gram-negative bacteria is thin (about five to 10 nm thick); in E. coli, the peptidoglycan is probably only a monolayer thick. Outside the peptidoglycan layer in the Gram-negative envelope is an outer membrane structure (well-nigh vii.v to 10 nm thick). In about Gram-negative bacteria, this membrane structure is anchored noncovalently to lipoprotein molecules (Braun'due south lipoprotein), which, in plough, are covalently linked to the peptidoglycan. The lipopolysaccharides of the Gram-negative jail cell envelope grade office of the outer leaflet of the outer membrane structure.
The system and overall dimensions of the outer membrane of the Gram-negative cell envelope are like to those of the plasma membrane (about 7.5 nm thick). Moreover, in Gram-negative bacteria such as E. coli, the outer and inner membranes adhere to each other at several hundred sites (Bayer patches); these sites can break up the continuity of the peptidoglycan layer. Table 2-ii summarizes the major classes of chemic constituents in the walls and envelopes of Gram-positive and Gram-negative leaner.
Table two-two
Major Classes of Chemic Components in Bacterial Walls and Envelopes.
The bones differences in surface structures of Gram-positive and Gram-negative leaner explicate the results of Gram staining. Both Gram-positive and Gram-negative leaner take upward the aforementioned amounts of crystal violet (CV) and iodine (I). The CV-I complex, still, is trapped inside the Gram-positive jail cell by the aridity and reduced porosity of the thick cell wall every bit a event of the differential washing footstep with 95 percentage ethanol or other solvent mixture. In contrast, the thin peptidoglycan layer and probable discontinuities at the membrane adhesion sites do not impede solvent extraction of the CV-I complex from the Gram-negative jail cell. The above mechanism of the Gram stain based on the structural differences betwixt the two groups has been confirmed by sophisticated methods of electron microscopy (come across Ref. Bereridge and Daries). The sequence of steps in the Gram stain differentiation is illustrated diagrammatically in Figure 2-7. Moreover, mechanical disruption of the prison cell wall of Gram-positive organisms or its enzymatic removal with lysozyme results in complete extraction of the CV-I circuitous and conversion to a Gram-negative reaction. Therefore, autolytic wall-degrading enzymes that crusade jail cell wall breakage may account for Gram-negative or variable reactions in cultures of Gram-positive organisms (such equally Staphylococcus aureus, Clostridium perfringens, Corynebacterium diphtheriae, and some Bacillus spp.).

Figure 2-7
General sequence of steps in the Gram stain procedure and the resultant staining of Gram-positive and Gram-negative bacteria.
Peptidoglycan
Unique features of almost all prokaryotic cells (except for Halobacterium halobium and mycoplasmas) are prison cell wall peptidoglycan and the specific enzymes involved in its biosynthesis. These enzymes are target sites for inhibition of peptidoglycan synthesis by specific antibiotics. The primary chemic structures of peptidoglycans of both Gram-positive and Gram-negative bacteria accept been established; they consist of a glycan courage of repeating groups of β1, 4-linked disaccharides of β1,4-Due north-acetylmuramyl-N-acetylglucosamine. Tetrapeptides of L-alanine-D-isoglutamic acid-L-lysine (or diaminopimelic acid)-n-alanine are linked through the carboxyl group by amide linkage of muramic acid residues of the glycan chains; the D-alanine residues are directly cross-linked to the 𝛆-amino group of lysine or diaminopimelic acid on a neighboring tetrapeptide, or they are linked by a peptide bridge. In S. aureus peptidoglycan, a glycine pentapeptide span links the ii adjacent peptide structures. The extent of directly or peptide-bridge cross-linking varies from one peptidoglycan to some other. The staphylococcal peptidoglycan is highly cantankerous-linked, whereas that of E. coli is much less and so, and has a more open peptidoglycan mesh. The diamino acid providing the 𝛆-amino group for cross-linking is lysine or diaminopimelic acid, the latter being uniformly present in Gram-negative peptidoglycans. The construction of the peptidoglycan is illustrated in Effigy 2-8. A peptidoglycan with a chemical structure substantially dissimilar from that of all eubacteria has been discovered in certain archaebacteria. Instead of muramic acid, this peptidoglycan contains talosaminuronic acrid and lacks the D-amino acids found in the eubacterial peptidoglycans. Interestingly, organisms containing this wall polymer (referred to as pseudomurein) are insensitive to penicillin, an inhibitor of the transpeptidases involved in peptidoglycan biosynthesis in eubacteria.
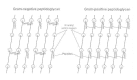
Figure ii-viii
Diagrammatic representation of peptidoglycan structures with adjacent glycan strands cross-linked directly from the carboxyterminal D-alanine to the 𝛆-amino group of an adjacent tetrapeptide or through a peptide cross bridge, N-acetylmuramic acrid, (more...)
The ß-ane,4 glycosidic bond between N-acetylmuramic acid and N-acetylglucosamine is specifically broken by the bacteriolytic enzyme lysozyme. Widely distributed in nature, this enzyme is present in human tissues and secretions and can cause complete digestion of the peptidoglycan walls of sensitive organisms. When lysozyme is allowed to digest the cell wall of Gram-positive leaner suspended in an osmotic stabilizer (such equally sucrose), protoplasts are formed. These protoplasts are able to survive and continue to grow on suitable media in the wall-less land. Gram-negative bacteria treated similarly produce spheroplasts, which retain much of the outer membrane construction. The dependence of bacterial shape on the peptidoglycan is shown by the transformation of rod-shaped bacteria to spherical protoplasts (spheroplasts) after enzymatic breakdown of the peptidoglycan. The mechanical protection afforded by the wall peptidoglycan layer is evident in the osmotic fragility of both protoplasts and spheroplasts. There are ii groups of bacteria that lack the protective jail cell wall peptidoglycan construction, the Mycoplasma species, 1 of which causes atypical pneumonia and some genitourinary tract infections and the L-forms, which originate from Gram-positive or Gram-negative bacteria and are and then designated because of their discovery and clarification at the Lister Institute, London. The mycoplasmas and L-forms are all Gram-negative and insensitive to penicillin and are divisional by a surface membrane structure. L-forms arising "spontaneously" in cultures or isolated from infections are structurally related to protoplasts and spheroplasts; all iii forms (protoplasts, spheroplasts, and L-forms) revert infrequently and just nether special weather condition.
Teichoic Acids
Wall teichoic acids are establish only in certain Gram-positive bacteria (such as staphylococci, streptococci, lactobacilli, and Bacillus spp.); so far, they have non been found in gram- negative organisms. Teichoic acids are polyol phosphate polymers, with either ribitol or glycerol linked by phosphodiester bonds; their structures are illustrated in Figure 2-nine. Substituent groups on the polyol bondage can include D-alanine (ester linked), N-acetylglucosamine, N-acetylgalactosamine, and glucose; the substituent is characteristic for the teichoic acid from a item bacterial species and can act as a specific antigenic determinant. Teichoic acids are covalently linked to the peptidoglycan. These highly negatively charged polymers of the bacterial wall tin can serve every bit a cation-sequestering machinery.
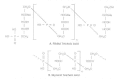
Figure 2-9
Structures of prison cell wall teichoic acids. (A) Ribitol teichoic acid with repeating units of 1,5-phosphodiester linkages of D-ribitol and D-alanyl ester on position two and glycosyl substituents (R) on position 4. The glycosyl groups may abe N-acetylglucosaminyl (more...)
Accessory Wall Polymers
In addition to the principal cell wall polymers, the walls of sure Gram-positive bacteria possess polysaccharide molecules linked to the peptidoglycan. For example, the C polysaccharide of streptococci confers group specificity. Acidic polysaccharides attached to the peptidoglycan are called teichuronic acids. Mycobacteria take peptidoglycolipids, glycolipids, and waxes associated with the jail cell wall.
Lipopolysaccharides
A characteristic feature of Gram-negative bacteria is possession of various types of circuitous macromolecular lipopolysaccharide (LPS). And so far, only i Gram-positive organism, Listeria monocytogenes, has been found to incorporate an authentic LPS. The LPS of this bacterium and those of all Gram-negative species are also called endotoxins, thereby distinguishing these cell-bound, heat-stable toxins from estrus-labile, protein exotoxins secreted into culture media. Endotoxins possess an array of powerful biologic activities and play an of import role in the pathogenesis of many Gram-negative bacterial infections. In add-on to causing endotoxic shock, LPS is pyrogenic, can actuate macrophages and complement, is mitogenic for B lymphocytes, induces interferon production, causes tissue necrosis and tumor regression, and has adjuvant properties. The endotoxic properties of LPS reside largely in the lipid A components. Usually, the LPS molecules have iii regions: the lipid A construction required for insertion in the outer leaflet of the outer membrane bilayer; a covalently attached cadre composed of two-keto-3deoxyoctonic acid (KDO), heptose, ethanolamine, North-acetylglucosamine, glucose, and galactose; and polysaccharide chains linked to the core. The polysaccharide chains plant the O-antigens of the Gram-negative bacteria, and the individual monosaccharide constituents confer serologic specificity on these components. Figure ii-x depicts the structure of LPS. Although it has been known that lipid A is equanimous of β1,half-dozen-linked D-glucosamine disaccharide substituted with phosphomonester groups at positions 4' and i, uncertainties have existed about the attachment positions of the six fatty acid acyl and KDO groups on the disaccharide. The demonstration of the construction of lipid A of LPS of a heptoseless mutant of Salmonella typhimurium has established that amide-linked hydroxymyristoyl and lauroxymyristoyl groups are fastened to the nitrogen of the 2- and 2'-carbons, respectively, and that hydroxymyristoyl and myristoxymyristoyl groups are fastened to the oxygen of the three- and 3'-carbons of the disaccharide, respectively. Therefore, only position 6' is left for zipper of KDO units.
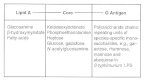
Figure two-10
The iii major, covalently linked regions that form the typical LPS.
LPS and phospholipids help confer disproportion to the outer membrane of the Gram-negative bacteria, with the hydrophilic polysaccharide chains outermost. Each LPS is held in the outer membrane by relatively weak cohesive forces (ionic and hydrophobic interactions) and can be dissociated from the prison cell surface with surface-agile agents.
As in peptidoglycan biosynthesis, LPS molecules are assembled at the plasma or inner membrane. These newly formed molecules are initially inserted into the outer-inner membrane adhesion sites.
Outer Membrane of Gram-Negative Bacteria
In thin sections, the outer membranes of Gram-negative bacteria appear broadly similar to the plasma or inner membranes; however, they differ from the inner membranes and walls of Gram-positive bacteria in numerous respects. The lipid A of LPS is inserted with phospholipids to create the outer leaflet of the bilayer structure; the lipid portion of the lipoprotein and phospholipid grade the inner leaflet of the outer membrane bilayer of most Gram-negative leaner (Fig. 2-6).
In addition to these components, the outer membrane possesses several major outer membrane proteins; the virtually arable is called porin. The assembled subunits of porin form a channel that limits the passage of hydrophilic molecules across the outer membrane barrier to those having molecular weights that are usually less than 600 to 700. Bear witness besides suggests that hydrophobic pathways exist beyond the outer membrane and are partly responsible for the differential penetration and effectiveness of certain b-lactam antibiotics (ampicillin, cephalosporins) that are active against various Gram-negative bacteria. Although the outer membranes deed as a permeability barrier or molecular sieve, they exercise non announced to possess free energy-transducing systems to drive active transport. Several outer membrane proteins, however, are involved in the specific uptake of metabolites (maltose, vitamin B12, nucleosides) and iron from the medium. Thus, outer membranes of the Gram-negative bacteria provide a selective barrier to external molecules and thereby forbid the loss of metabolite-binding proteins and hydrolytic enzymes (nucleases, alkali metal phosphatase) found in the periplasmic infinite. The periplasmic space is the region between the outer surface of the inner (plasma) membrane and the inner surface of the outer membrane (Figure 2-6). Thus, Gram-negative bacteria accept a cellular compartment that has no equivalent in Gram-positive organisms. In addition to the hydrolytic enzymes, the periplasmic infinite holds binding proteins (proteins that specifically bind sugars, amino acids, and inorganic ions) involved in membrane transport and chemotactic receptor activities. Moreover, plasmid-encoded b-lactamases and aminoglycoside-modifying enzymes (phosphorylation or adenylation) in the periplasmic infinite produce antibiotic resistance by degrading or modifying an antibiotic in transit to its target sites on the membrane (penicillin-binding proteins) or on the ribosomes (aminoglycosides). These periplasmic proteins can be released past subjecting the cells to osmotic stupor and after treatment with the chelating agent ethylenediaminetetraacetic acid.
Intracellular Components
Plasma (Cytoplasmic) Membranes
Bacterial plasma membranes, the functional equivalents of eukaryotic plasma membranes, are referred to variously every bit cytoplasmic, protoplast, or (in Gram-negative organisms) inner membranes. Like in overall dimensions and appearance in thin sections to biomembranes from eukaryotic cells, they are composed primarily of proteins and lipids (principally phospholipids). Protein-to-lipid ratios of bacterial plasma membranes are approximately 3: 1, shut to those for mitochondrial membranes. Dissimilar eukaryotic jail cell membranes, the bacterial membrane (except for Mycoplasma species and certain methylotrophic leaner) has no sterols, and bacteria lack the enzymes required for sterol biosynthesis.
Although their composition is like to that of inner membranes of Gram-negative species, cytoplasmic membranes from Gram-positive bacteria possess a form of macromolecules not present in the Gram-negative membranes. Many Gram-positive bacterial membranes comprise membrane-bound lipoteichoic acid, and species lacking this component (such as Micrococcus and Sarcina spp.) contain an analogous membrane-spring succinylated lipomannan. Lipoteichoic acids are structurally like to the cell wall glycerol teichoic acids in that they have basal polyglycerol phosphodiester 1-3 linked chains (Fig. 2-nine). These chains terminate with the phosphomonoester end of the polymer, which is linked covalently to either a glycolipid or a phosphatidyl glycolipid moiety. Thus, a hydrophobic tail is provided for anchoring in the membrane lipid layers (Fig. 2-6A). As in the cell wall glycerol teichoic acrid, the lipoteichoic acids can have glycosidic and D-alanyl ester substituents on the C-two position of the glycerol.
Both membrane-jump lipoteichoic acid and membrane-spring succinylated lipomannan can be detected as antigens on the jail cell surface, and the glycerol-phosphate and succinylated mannan chains appear to extend through the jail cell wall structure (Fig. two-half dozen). This class of polymer has non however been found in the cytoplasmic membranes of Gram-negative organisms. In both instances, the lipoteichoic acids and the lipomannans are negatively charged components and tin can sequester positively charged substances. They take been implicated in adhesion to host cells, but their functions remain to exist elucidated.
Multiple functions are performed by the plasma membranes of both Gram-positive and Gram-negative bacteria. Plasma membranes are the site of active transport, respiratory concatenation components, energy-transducing systems, the H+-ATPase of the proton pump (run across Chapter 4), and membrane stages in the biosynthesis of phospholipids, peptidoglycan, LPS, and capsular polysaccharides. In essence, the bacterial cytoplasmic membrane is a multifunction structure that combines the mitochondrial send and biosynthetic functions that are usually compartmentalized in discrete bleary organelles in eukaryotic cells. The plasma membrane is also the anchoring site for DNA and provides the jail cell with a machinery (every bit withal unknown) for separation of sister chromosomes.
Mesosomes
Thin sections of Gram-positive bacteria reveal the presence of vesicular or tubular-vesicular membrane structures called mesosomes, which are evidently formed by an invagination of the plasma membrane. These structures are much more prominent in Gram-positive than in Gram-negative organisms. At one fourth dimension, the mesosomal vesicles were thought to be equivalent to bacterial mitochondria; withal, many other membrane functions take also been attributed to the mesosomes. At nowadays, there is no satisfactory show to suggest that they have a unique biochemical or physiologic function. Indeed, electron-microscopic studies accept suggested that the mesosomes, every bit usually seen in thin sections, may arise from membrane perturbation and fixation artifacts. No full general understanding exists virtually this theory, however, and some evidence indicates that mesosomes may be related to events in the cell sectionalisation cycle.
Other Intracellular Components
In addition to the nucleoid and cytoplasm (cytosol), the intracellular compartment of the bacterial cell is densely packed with ribosomes of the 70S type (Fig. two-2). These ribonucleoprotein particles, which have a diameter of 18 nm, are not bundled on a membranous rough endoplasmic reticulum as they are in eukaryotic cells. Other granular inclusions randomly distributed in the cytoplasm of various species include metabolic reserve particles such as poly-β-hydroxybutyrate (PHB), polysaccharide and glycogen-like granules, and polymetaphosphate or metachromatic granules.
Endospores are highly heat-resistant, dehydrated resting cells formed intracellularly in members of the genera Bacillus and Clostridium. Sporulation, the procedure of forming endospores, is an unusual holding of sure bacteria. The serial of biochemical and morphologic changes that occur during sporulation represent true differentiation within the bike of the bacterial prison cell. The process, which usually begins in the stationary phase of the vegetative cell cycle, is initiated past depletion of nutrients (usually readily utilizable sources of carbon or nitrogen, or both). The cell then undergoes a highly complex, well-divers sequence of morphologic and biochemical events that ultimately lead to the germination of mature endospores. As many equally seven distinct stages have been recognized by morphologic and biochemical studies of sporulating Bacillus species: stage 0, vegetative cells with two chromosomes at the terminate of exponential growth; stage I, germination of axial chromatin filament and excretion of exoenzymes, including proteases; stage 2, forespore septum formation and segregation of nuclear material into two compartments; stage Three, spore protoplast germination and meridian of tricarboxylic acid and glyoxylate bike enzyme levels; phase Iv, cortex formation and refractile appearance of spore; stage V, spore coat protein formation; stage VI, spore maturation, modification of cortical peptidoglycan, uptake of dipicolinic acid (a unique endospore product) and calcium, and development of resistance to heat and organic solvents; and stage 7, final maturation and liberation of endospores from mother cells (in some species).
When newly formed, endospores appear as circular, highly refractile cells inside the vegetative jail cell wall, or sporangium. Some strains produce autolysins that digest the walls and liberate free endospores. The spore protoplast, or cadre, contains a complete nucleus, ribosomes, and energy generating components that are enclosed within a modified cytoplasmic membrane. The peptidoglycan spore wall surrounds the spore membrane; on germination, this wall becomes the vegetative cell wall. Surrounding the spore wall is a thick cortex that contains an unusual type of peptidoglycan, which is rapidly released on germination. A spore coat of keratinlike poly peptide encases the spore contained inside a membrane (the exosporium). During maturation, the spore protoplast dehydrates and the spore becomes refractile and resistant to estrus, radiations, pressure, desiccation, and chemicals; these properties correlate with the cortical peptidoglycan and the presence of big amounts of calcium dipicolinate.
Recent bear witness indicated that the spores of Bacillus spharicus were revived which had been preserved in amber for more than than 25 million years. Their claims demand to be reevaluated. Figure 2-11 illustrates the principal structural features of a typical endospore (Bacillus megaterium) on initiation of the germination procedure. The thin section of the spore shows the ruptured, thick spore coat and the cortex surrounding the spore protoplast with the germinal cell wall that becomes the vegetative wall on outgrowth.
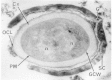
Effigy 2-eleven
Electron micrograph of a thin section of a Bacillus megaterium spore showing the thick spore coat (SC), germinal groove (G) in the spore coat, outer cortex layer (OCL) and cortex (Cx) germinal prison cell wall layer (GCW), underlying spore protoplast membrane (more...)
References
-
Beveridge TJ, Davies JA. Cellular responses of Bacillus subtilis and Escherichia coli to the Gram stain. J Bacteriol. 1983;156:846. [PMC free article: PMC217903] [PubMed: 6195148]
-
Costerton JW, Ingram JM, Cheng KJ. Construction and function of the cell envelope of gram-negative leaner. Bacteriol Rev. 1974;38:87. [PMC gratis article: PMC413842] [PubMed: 4601163]
-
Ghuysen J-M, Hakenbeck R: Bacterial jail cell wall. Elsevier, 1994 .
-
Gould GW, Hurst A (eds): The Bacterial Spore. Academic Press, San Diego, 1969 .
-
Jawetz E, Melnick JL, Adelberg EA: Medical Microbiology. Appleton & Lange, East Norwalk, CT, 1989 .
-
Rogers HJ: Bacterial Prison cell Structure. American Society for Microbiology, Washington, D.C., 1983 .
-
Wright A, Tipper DJ: The outer membrane of gram-negative bacteria. p. 427. In Sokatch JR, Ornston LN (eds): The bacteria. Vol. 7. Bookish Press, San Diego, 1979 .
Copyright © 1996, The Academy of Texas Medical Branch at Galveston.
Bookshelf ID: NBK8477 PMID: 21413343
Source: https://www.ncbi.nlm.nih.gov/books/NBK8477/
0 Response to "All of the Following Can Form Filaments Except __________."
Post a Comment